3.7. Comparison of MD vs. Conformer Generators¶
To assess how well the chemical informatics-based conformer generators perform compared to the accelerated MD simulations, we performed various runs of the RDKit and OMEGA conformer generators with different input parameters. An overlay of the RDKit and OMEGA structures onto the dPCA derived PES plots is shown in Fig. 3.19 for Omega, and Fig. 3.20 for RDKit. Depending on the parameter choice, we were able to increase sampling for both conformer generators. Generally, the two conformer generators reproduced most of the in GaMD observed structures, but the RDKit conformer generator was unable to find the global GaMD minimum. At least visually, RDKit and OMEGA performed comparably otherwise. This only considers the backbone structures as we compared to the dPCA representation of the GaMD simulation. There was no clustering or higher density around PES minima observed: RDKit and Omega seem to produce geometrically plausible structures, but no information is provided about which are most relevant. Also, the conformer generators do not take explicit solvent effects into account.
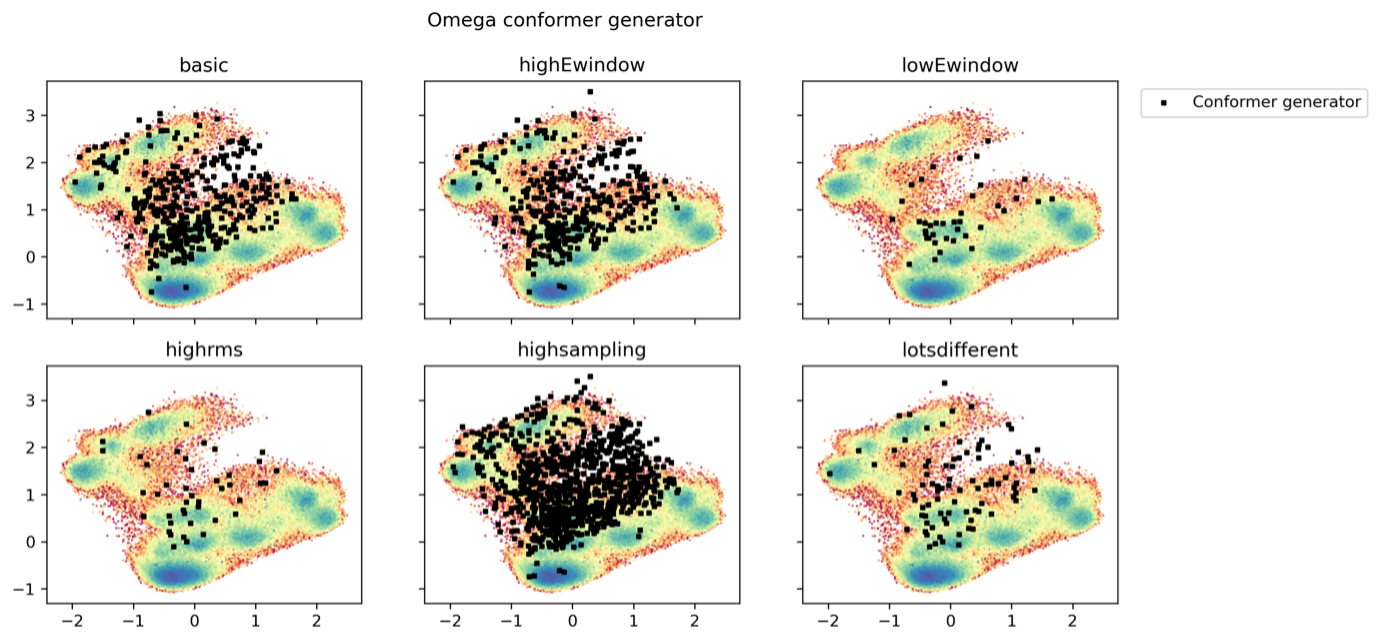
Fig. 3.19 Different Omega-Macrocycle conformer generator parameters overlayed on the 2000 ns GaMD simulation in \(\rm H_{2}O\)¶
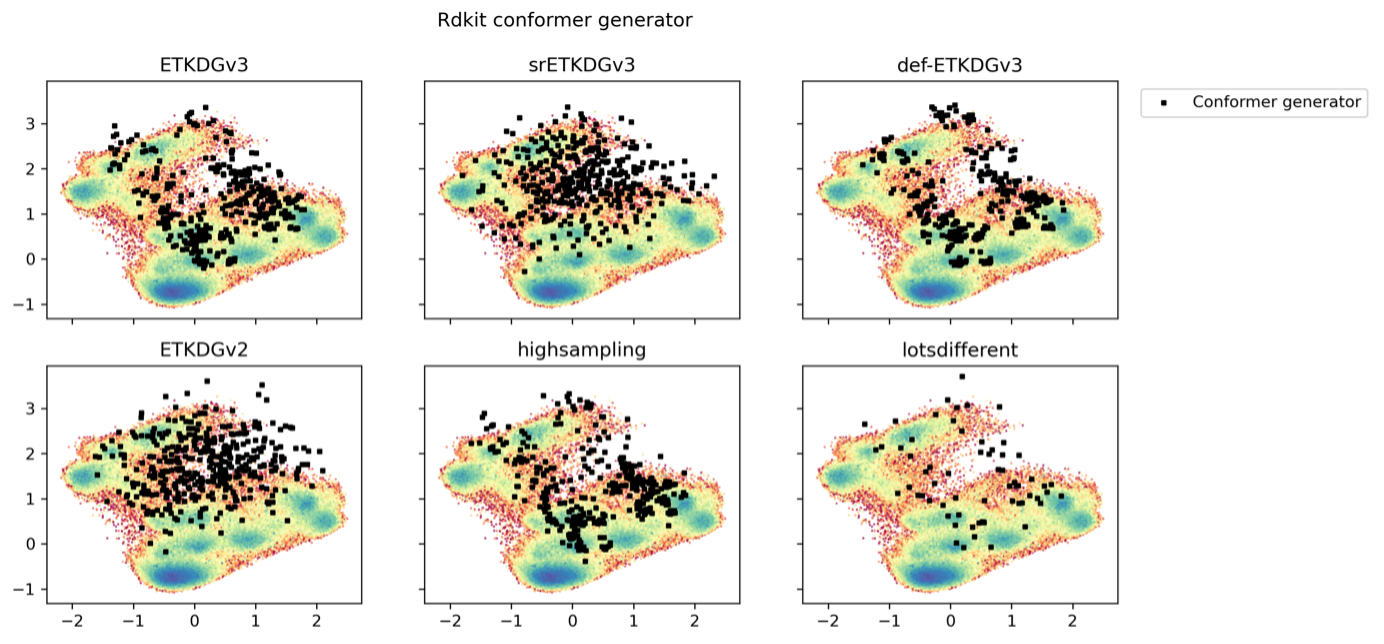
Fig. 3.20 Different RDKit conformer generators overlayed on the 2000 ns GaMD simulation in \(\rm H_{2}O\).¶
